Chapter 3 Reading the Excel data file
3.1 TBA How much finished
90%
3.2 Introduction
In the previous chapter I described the format of the Excel file R/quick-start/rocCr.xlsx corresponding to a small factorial ROC dataset. Described here is how to read this file in order to create an RJafroc dataset. It introduces the RJafroc function DfReadDataFile(). Also shown are the correspondences between values in the Excel file and the dataset object.
3.3 The structure of an ROC dataset
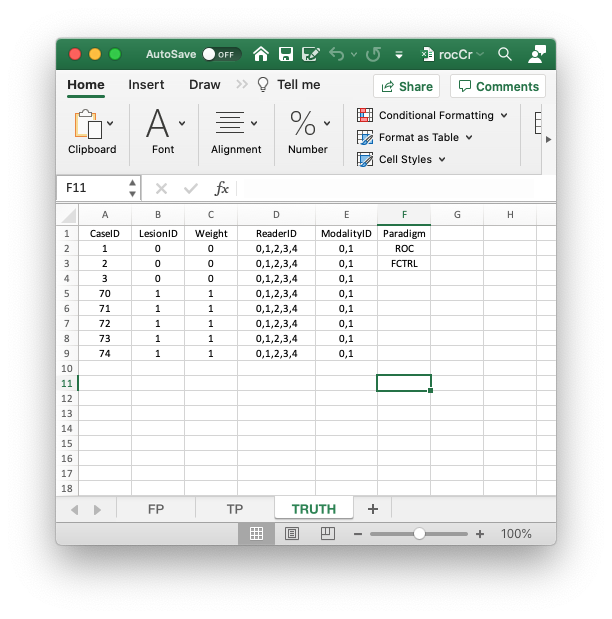
In the following code chunk the second statement reads the Excel file using the function DfReadDataFile() and saves it to object x. The third statement shows the structure of x.
rocCr <- "R/quick-start/rocCr.xlsx"
x <- DfReadDataFile(rocCr, newExcelFileFormat = TRUE)
str(x)
#> List of 3
#> $ ratings :List of 3
#> ..$ NL : num [1:2, 1:5, 1:8, 1] 1 3 2 3 2 2 1 2 3 2 ...
#> ..$ LL : num [1:2, 1:5, 1:5, 1] 5 5 5 5 5 5 5 5 5 5 ...
#> ..$ LL_IL: logi NA
#> $ lesions :List of 3
#> ..$ perCase: int [1:5] 1 1 1 1 1
#> ..$ IDs : num [1:5, 1] 1 1 1 1 1
#> ..$ weights: num [1:5, 1] 1 1 1 1 1
#> $ descriptions:List of 7
#> ..$ fileName : chr "rocCr"
#> ..$ type : chr "ROC"
#> ..$ name : logi NA
#> ..$ truthTableStr: num [1:2, 1:5, 1:8, 1:2] 1 1 1 1 1 1 1 1 1 1 ...
#> ..$ design : chr "FCTRL"
#> ..$ modalityID : Named chr [1:2] "0" "1"
#> .. ..- attr(*, "names")= chr [1:2] "0" "1"
#> ..$ readerID : Named chr [1:5] "0" "1" "2" "3" ...
#> .. ..- attr(*, "names")= chr [1:5] "0" "1" "2" "3" ...- In the above code chunk flag
newExcelFileFormatis set toTRUEas otherwise columns D - F in theTruthworksheet are ignored and the dataset is assumed to be factorial, withdataType“automatically” determined from the contents of the FP and TP worksheets.3 - Flag
newExcelFileFormat = FALSE, the default, is for compatibility with older JAFROC format Excel files, which did not have columns D - F in theTruthworksheet. Its usage is deprecated. - The dataset object
xis alistvariable with 3 members:ratings,lesionsanddescriptions. - The
x$ratingsmember contains 3 sub-lists.- The
x$ratings$NLmember, with dimension [2, 5, 8, 1], contains the ratings of normal cases. The first dimension (2) is the number of treatments, the second (5) is the number of readers and the third (8) is the total number of cases. For ROC datasets the fourth dimension is always unity. The five extra values4 in the third dimension, which are filled withNAs, are needed for compatibility with FROC datasets. - The
x$ratings$LL, with dimension [2, 5, 5, 1], contains the ratings of abnormal cases. The third dimension (5) corresponds to the 5 diseased cases. - The
x$ratings$LL_ILmember, equal to NA’; this member is there for compatibility with LROC data,_ILdenotes incorrect-localizations.
- The
- The
x$lesionsmember contains 3 sub-lists.- The
x$lesions$perCasemember is a vector with 5 ones representing the 5 diseased cases in the dataset. - The
x$lesions$IDsmember is an array with 5 ones. - The
x$lesions$weightsmember is an array with 5 ones. - These are irrelevant for ROC datasets. They are there for compatibility with FROC datasets.
- The
- The
x$descriptionsmember contains 7 sub-lists.- The
x$descriptions$fileNamemember is the base name of the file that was read to create this dataset, “rocCr” in the current example, otherwise it isNA(the latter would apply, for example, for a simulated dataset). - The
x$descriptions$typemember indicates that this is anROCdataset. - The
x$descriptions$namemember is the name of this dataset, if it is an embedded dataset, otherwiseNA. - The
x$descriptions$truthTableStrmember, with dimension [2, 5, 8, 2], quantifies the structure of the dataset, as explained in TBA Vignette #3 (it is used to check for data entry errors). - The
x$descriptions$designmember specifies the dataset design, which is “FCTRL” in the present example (a factorial dataset). - The
x$descriptions$modalityIDmember is a vector with two elements"0"and"1", naming the two modalities. - The
x$readerIDmember is a vector with five elements"0","1","2","3"and"4", naming the five readers.
- The
3.4 Correspondence between NL member of dataset and the FP worksheet
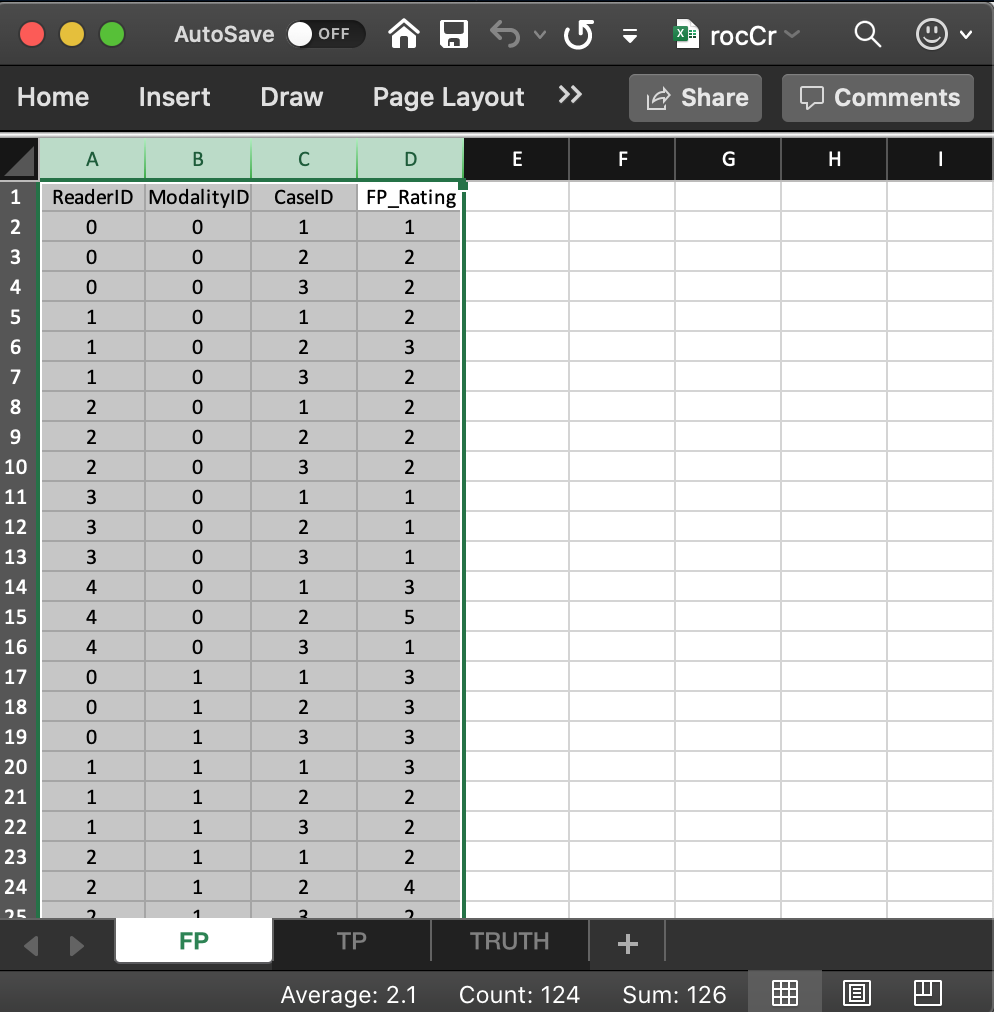
- The list member
x$ratings$NLis an array withdim = c(2,5,8,1).- The first dimension (2) comes from the number of modalities.
- The second dimension (5) comes from the number of readers.
- The third dimension (8) comes from the total number of cases.
- The fourth dimension is alway 1 for an ROC dataset.
- The value of
x$ratings$NL[1,5,2,1], i.e., 5, corresponds to row 15 of the FP table, i.e., toModalityID= 0,ReaderID= 4 andCaseID= 2. - The value of
x$ratings$NL[2,3,2,1], i.e., 4, corresponds to row 24 of the FP table, i.e., toModalityID1,ReaderID2 andCaseID2. - All values for case index > 3 and case index <= 8 are
-Inf. For example the value ofx$ratings$NL[2,3,4,1]is-Inf. This is because there are only 3 non-diseased cases. The extra length is needed for compatibility with FROC datasets.
3.5 Case-index vs. caseID
- Regardless of what order they occur in the worksheet, the non-diseased cases are always indexed first. In the current example the case indices are 1, 2 and 3, corresponding to the three non-diseased cases with
caseIDsequal to 1, 2 and 3. - Regardless of what order they occur in the worksheet, in the NL array the diseased cases are always indexed after the last non-diseased case. In the current example the case indices in the
NLarray are 4, 5, 6, 7 and 8, corresponding to the five diseased cases withcaseIDsequal to 70, 71, 72, 73, and 74. In theLLarray they are numbered 1, 2, 3, 4 and 5, corresponding to the five diseased cases withcaseIDsequal to 70, 71, 72, 73, and 74. Some examples follow: x$ratings$NL[1,3,2,1], a FP rating, refers toModalityID0,ReaderID2 andCaseID2 (since the modality and reader IDs start with 0).x$ratings$NL[2,5,4,1], a FP rating, refers toModalityID1,ReaderID4 andCaseID70, the first diseased case; this is-Inf.x$ratings$NL[1,4,8,1], a FP rating, refers toModalityID0,ReaderID3 andCaseID74, the last diseased case; this is-Inf.x$ratings$NL[1,3,9,1], a FP rating, is an illegal value, as the third index cannot exceed 8.x$ratings$NL[1,3,8,2], a FP rating, is an illegal value, as the fourth index cannot exceed 1 for an ROC dataset.x$ratings$LL[1,3,1,1], a TP rating, refers toModalityID0,ReaderID2 andCaseID70, the first diseased case.x$ratings$LL[2,5,4,1], a TP rating, refers toModalityID1,ReaderID4 andCaseID73, the fourth diseased case.
3.6 Correspondence between LL member of dataset and the TP worksheet
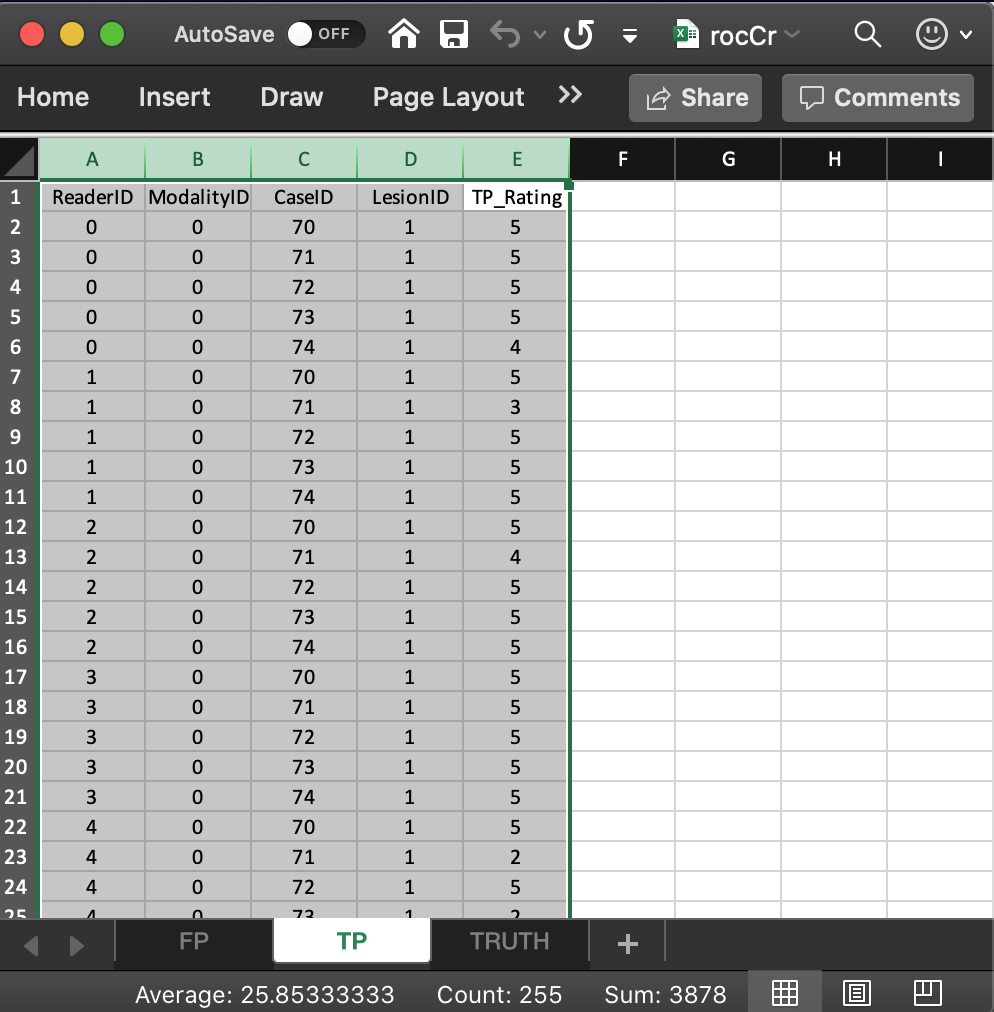
- The list member
x$ratings$LLis an array withdim = c(2,5,5,1).- The first dimension (2) comes from the number of modalities.
- The second dimension (5) comes from the number of readers.
- The third dimension (5) comes from the number of diseased cases.
- The fourth dimension is always 1 for an ROC dataset.
- The value of
x$ratings$LL[1,1,5,1], i.e., 4, corresponds to row 6 of the TP table, i.e., toModalityID= 0,ReaderID= 0 andCaseID= 74. - The value of
x$ratings$LL[1,2,2,1], i.e., 3, corresponds to row 8 of the TP table, i.e., toModalityID= 0,ReaderID= 1 andCaseID= 71. - The value of
x$ratings$LL[1,4,4,1], i.e., 5, corresponds to row 21 of the TP table, i.e., toModalityID= 0,ReaderID= 3 andCaseID= 74. - The value of
x$ratings$LL[1,5,2,1], i.e., 2, corresponds to row 23 of the TP table, i.e., toModalityID= 0,ReaderID= 4 andCaseID= 71. - There are no
-Infvalues inx$ratings$LL:any(x$ratings$LL == -Inf)= FALSE. This is true for any ROC dataset.