Chapter 4 Data format and reading LROC data
4.2 Introduction
In the Localization Receiver Operating Characteristic (LROC) paradigm (S. Starr et al. 1977; S. J. Starr et al. 1975; Swensson 1996) the observer is restricted to at most one mark-rating pair per case. Additionally, each diseased case has exactly one lesion. On a diseased case and if the mark is close to the real lesion the investigator classifies the mark as a correct-localization (CL). Otherwise it is classified as an incorrect-localization (IL). On a non-diseased case the mark is always classified as a false-positive (FP).
The paradigm is illustrated with a few toy data files, R/quick-start/lroc?.xlsx, where ? is 1 or 2. These files illustrate two-modality three-reader LROC datasets with 3 non-diseased and 5 diseased cases.
File lroc1.xlsx illustrates the classic (i.e., as originally introduced) LROC paradigm where one mark per case is forced.
File lroc2.xlsx illustrates the paradigm when one mark-rating pair per case is not forced. There is some history behind this: the basic issue was what was the observer supposed to do when there there was nothing to report? Swensson initially thought that even if there was nothing to report, there must be a region, selected from the set of very low confidence regions, which was most likely to be a lesion. Most radiologists had difficulty with the forced localization requirement - if they saw nothing suspicious, why should they be forced to indicate a location. The paradigm was subsequently altered so that if the confidence level was below a certain value, say 12 percent on a 0 to 100 scale, the radiologist did not have to report a location. LROCFIT software was modified accordingly, and internal to the software the mark was assigned a random location - which ended up being classified as an incorrect-localization in most cases. The fact that there are cases with nothing to report is accounted for in the radiological search model.
4.3 Truth worksheet
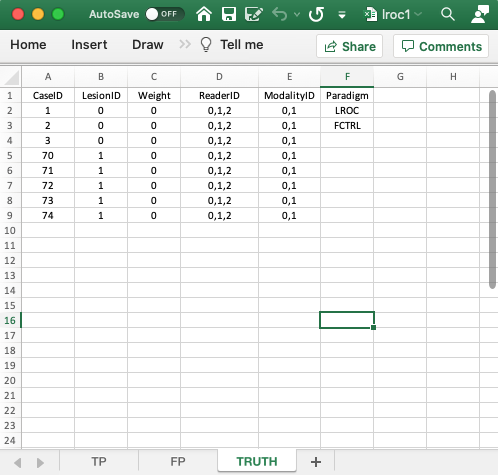
- The
Truthworksheet is similar to that described previously for the ROC and LROC paradigms. The only difference is the first entry in the Paradigm column, which isLROC. - Since a diseased case has one lesion, the first five columns contain as many rows as there are cases in the dataset. There being 8 cases in the dataset, there are 8 rows of data.
CaseID: unique integers representing the cases in the dataset: ‘1’, ‘2’, ‘3’, the 3 non-diseased cases, and ‘70’, ‘71’, ‘72’, ‘73’, ‘74’, the 5 diseased cases.
LesionID: integers 0 or 1.- Each 0 represents a non-diseased case,
- Each 1 represents the solitary lesion in the diseased case.
- There are 3 non-diseased cases in the dataset (the number of 0’s in the
LesionIDcolumn). - There are 5 diseased cases in the dataset (the number of 1’s in the
LesionIDcolumn). Weight: this column is filled with zeroes. With one lesion per case, the weights are irrelevant.ReaderID: In the example shown each cell has the value ‘0, 1, 2’. There are 3 readers in the dataset, labeled0,1and2.ModalityID: In the example each cell has the value0, 1. There are 2 modalities in the dataset, labeled0and1.Paradigm: The contents areLROCandFCTRL: this is anLROCdataset and the design is “factorial”.
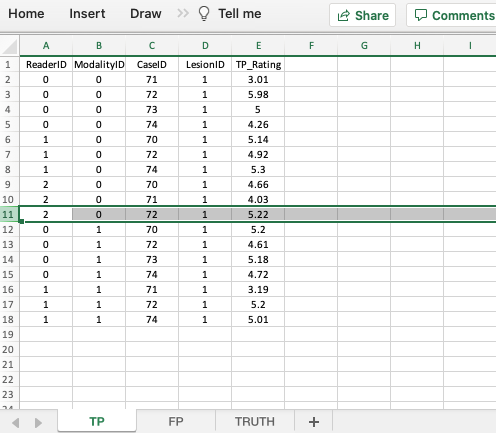
4.4 TP worksheet, forced localization true
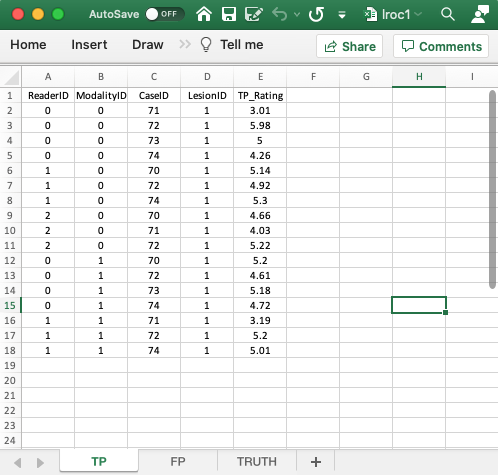
- The
TPworksheet is similar to that described previously for the ROC and FROC paradigms. - This worksheet can only have diseased cases. The presence of a non-diseased case in this worksheet will generate an error.
- The key difference is that for each modality-reader and diseased-case combination there can be at most one entry. Also, if a particular combination is missing in the TP worksheet then it must appear in the FP worksheet. This is because this is a forced-mark-per-case dataset.
- There can be at most 30 rows of data in this worksheet: 2 modalities times 3 readers times 5 diseased cases. Since there in fact only 17 rows of data, the missing 13 rows must occur in the FP worksheet.
- Recall that each entry in the TP worksheet represents a correct localization while each missing entry represents an incorrect localization. The incorrect localizations are recorded in the FP worksheet.
4.5 FP worksheet, forced localization true
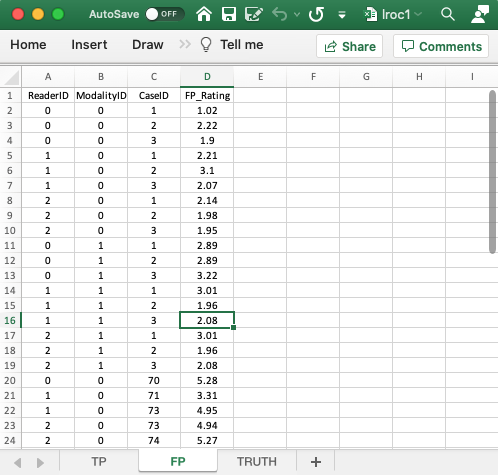
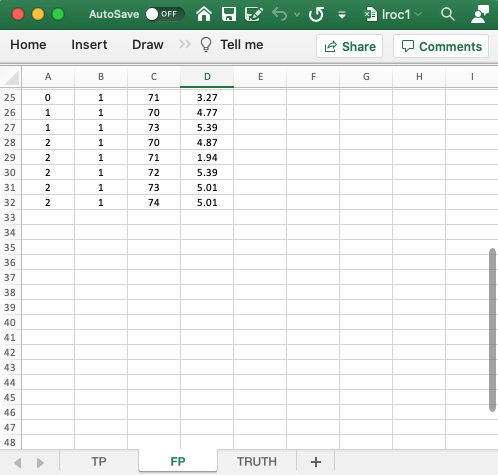
- The
FPworksheet is similar to that described previously for the ROC and FROC paradigms. - Because of the forced mark requirement, there are 18 rows of data corresponding to non-diseased cases: 2 modalities times 3 readers times 3 non-diseased cases. The missing 13 rows from the TP worksheet are listed next; these correspond to the incorrect localizations on diseased cases. Therefore, the total number of rows in this worksheet is 18 + 13 = 31.
- As an example, it is seen that for
modalityID= 0 andreaderID= 0,caseID= 70 does not appear in the TP worksheet. The lesion on this case was not localized; therefore it must appear in the FP worksheet as an incorrect localization, which is seen to be true in the FP worksheet. - As another example, for
modalityID= 0 andreaderID= 1,caseID= 71 does not appear in the TP worksheet; instead it appears in the FP worksheet. - As a final example, for
modalityID= 1 andreaderID= 2, none of the diseased cases appears in the TP worksheet; instead they all appear in the FP worksheet.
4.6 Reading forced localization true LROC dataset
The images shown above correspond to file R/quick-start/lroc1.xlsx. The next code reads this file into an R object x1. Note the usage of the lrocForcedMark flag, which is set to TRUE, because this is a forced localization LROC dataset.
lroc1 <- "R/quick-start/lroc1.xlsx"
x1 <- DfReadDataFile(lroc1, newExcelFileFormat = TRUE, lrocForcedMark = T)
str(x1)
#> List of 3
#> $ ratings :List of 3
#> ..$ NL : num [1:2, 1:3, 1:8, 1] 1.02 2.89 2.21 3.01 2.14 3.01 2.22 2.89 3.1 1.96 ...
#> ..$ LL : num [1:2, 1:3, 1:5, 1] -Inf 5.2 5.14 -Inf 4.66 ...
#> ..$ LL_IL: num [1:2, 1:3, 1:5, 1] 5.28 -Inf -Inf 4.77 -Inf ...
#> $ lesions :List of 3
#> ..$ perCase: int [1:5] 1 1 1 1 1
#> ..$ IDs : num [1:5, 1] 1 1 1 1 1
#> ..$ weights: num [1:5, 1] 1 1 1 1 1
#> $ descriptions:List of 7
#> ..$ fileName : chr "lroc1"
#> ..$ type : chr "LROC"
#> ..$ name : logi NA
#> ..$ truthTableStr: num [1:2, 1:3, 1:8, 1:2] 1 1 1 1 1 1 1 1 1 1 ...
#> ..$ design : chr "FCTRL"
#> ..$ modalityID : Named chr [1:2] "0" "1"
#> .. ..- attr(*, "names")= chr [1:2] "0" "1"
#> ..$ readerID : Named chr [1:3] "0" "1" "2"
#> .. ..- attr(*, "names")= chr [1:3] "0" "1" "2"This follows the general description in Chapter 2. The differences are described below.
x1$ratings$NLis a [2,3,8,1] dimension vector. For each modality and reader, only the first three elements, corresponding to the three non-diseased cases, are finite, the rest are-Inf.
For example:
x1$ratings$NL[1,1,,1]
#> [1] 1.02 2.22 1.90 -Inf -Inf -Inf -Inf -Inf- x1\(ratings\)LL
is a [2,3,5,1] dimension vector. For each modality and reader, only the first three elements, corresponding to the three non-diseased cases, are finite, the rest are-Inf`.
For example, since none of the lesions are localized for modalityID = 1 (second modality) and readerID = 2 (third reader), the following code yields a vector consisting of five -Inf values:
x1$ratings$LL[2,3,,1]
#> [1] -Inf -Inf -Inf -Inf -Infx1$ratings$LL_ILis a [2,3,5,1] dimension vector. These contain the ratings of incorrect localizations on diseased cases. For the just preceding modality-reader combination, this yields a vector with 5 finite values, the ratings of incorrect localizations formodalityID= 1 andreaderID= 2.
x1$ratings$LL_IL[2,3,,1]
#> [1] 4.87 1.94 5.39 5.01 5.014.8 FP worksheet, forced localization false
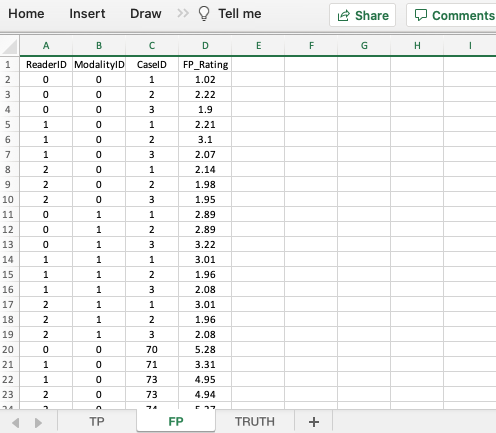
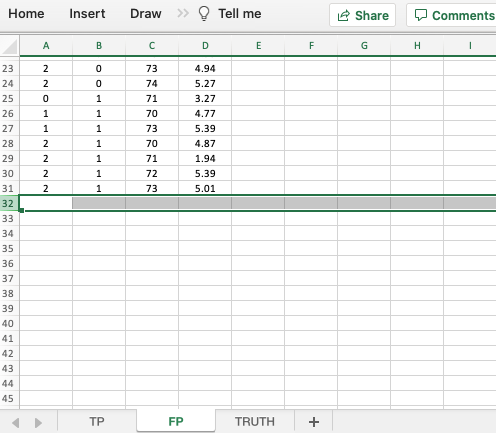
- If a particular modality-reader-case combination is missing in the TP worksheet then it need not appear in the FP worksheet. This is because this is not a forced-mark-per-case dataset.
- As an example,
modalityID= 1,readerID= 2 andcaseID= 74 does not appear in either TP or FP worksheets.
4.9 Reading forced localization false LROC dataset
The next example is for file R/quick-start/lroc2.xlsx. The following code reads this file into an R object x2. Note that for this dataset one must set the lrocForcedMark flag to FALSE, because this is not a forced localization LROC dataset. Setting lrocForcedMark flag to TRUE will generate an error.
lroc2 <- "R/quick-start/lroc2.xlsx"
x2 <- DfReadDataFile(lroc2, newExcelFileFormat = TRUE, lrocForcedMark = F)
str(x2)
#> List of 3
#> $ ratings :List of 3
#> ..$ NL : num [1:2, 1:3, 1:8, 1] 1.02 2.89 2.21 3.01 2.14 3.01 2.22 2.89 3.1 1.96 ...
#> ..$ LL : num [1:2, 1:3, 1:5, 1] -Inf 5.2 5.14 -Inf 4.66 ...
#> ..$ LL_IL: num [1:2, 1:3, 1:5, 1] 5.28 -Inf -Inf 4.77 -Inf ...
#> $ lesions :List of 3
#> ..$ perCase: int [1:5] 1 1 1 1 1
#> ..$ IDs : num [1:5, 1] 1 1 1 1 1
#> ..$ weights: num [1:5, 1] 1 1 1 1 1
#> $ descriptions:List of 7
#> ..$ fileName : chr "lroc2"
#> ..$ type : chr "LROC"
#> ..$ name : logi NA
#> ..$ truthTableStr: num [1:2, 1:3, 1:8, 1:2] 1 1 1 1 1 1 1 1 1 1 ...
#> ..$ design : chr "FCTRL"
#> ..$ modalityID : Named chr [1:2] "0" "1"
#> .. ..- attr(*, "names")= chr [1:2] "0" "1"
#> ..$ readerID : Named chr [1:3] "0" "1" "2"
#> .. ..- attr(*, "names")= chr [1:3] "0" "1" "2"- The
x2$ratings$LLarray is a [2,3,5,1] dimension vector. For each modality and reader, only the first three elements, corresponding to the three non-diseased cases, are finite, the rest are-Inf.
For example, since none of the lesions are localized for modalityID = 1 (second modality) and readerID = 2 (third reader), the following code yields a vector consisting of five -Inf values:
x2$ratings$LL[2,3,,1]
#> [1] -Inf -Inf -Inf -Inf -Inf- The
x2$ratings$LL_ILis a [2,3,5,1] dimension vector. These contain the ratings of incorrect localizations on diseased cases. For the just preceding modality-reader combination, this yields a vector with 4 finite values, the ratings of incorrect localizations formodalityID= 1 andreaderID= 2.
x2$ratings$LL_IL[2,3,,1]
#> [1] 4.87 1.94 5.39 5.01 -InfFor this modality-reader combination case 74 (i.e., the fifth diseased case) was unmarked. It does not appear in either the TP or the FP worksheet.
4.10 Summary
The difference from the previous data structures is the existence of LL_IL in the ratings list, which contains the ratings of incorrect localizations. Recall that for ROC and FROC paradigms this member was NA. When the data obeys forced localization, the corresponding flag should be set to TRUE, otherwise it should be set to FALSE. The default value of this flag is NA, which will work for ROC or FROC datasets. For LROC datasets it should be set to T/F.